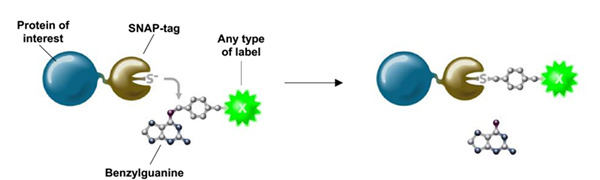
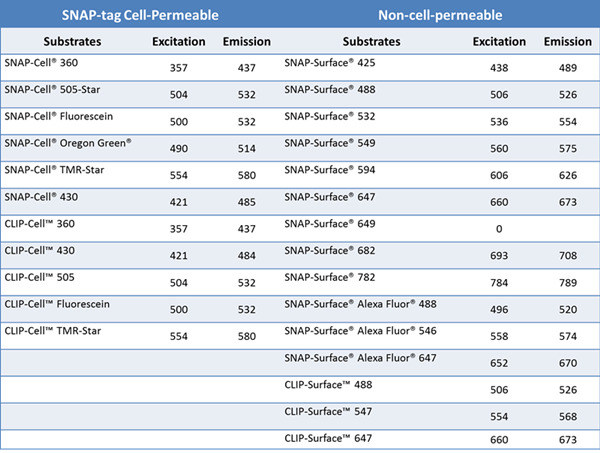
Author: Dr. Zhang Kun Leica Microsystems Life Science Applications Specialist Fluorescence microscopy plays an indispensable role in studying the life, activity and dynamics of protein molecules in living cells. Fluorescent proteins such as green fluorescent protein and the target protein are fused and expressed, and then the specific fluorescence emitted by the fluorescent protein is used to observe and trace the target protein molecule, which has been widely used in scientific research. However, fluorescent proteins have the disadvantages of low quantum yield, limited rate of maturation, easy exposure to environmental factors and easy formation of aggregates, which can cause artifacts in some special microscopic fluorescence imaging and affect the final experimental results. Therefore, scientists are also looking for and inventing more and more suitable living cell labeling methods to more accurately observe and record various microscopic life activities with more and more accurate microscopic imaging (such as ultra-high resolution). . This paper focuses on the characteristics and applications of novel labeling methods and dyes such as SNAP-tag and SiR. Introduction to SNAP-tag technology Figure 1. Schematic of the SNAP-tag tag. The labeling method has the advantages of high specificity, high speed, high stability and flexible selection of fluorescent probes. How can I label a protein of interest in a living cell with a specific fluorescent signal? When it comes to specificity, scientists first consider the interaction between enzymes and substrates. Therefore, starting from the enzymatic reaction, the Kai Johnsson research team at the Lausanne Institute of Technology in Switzerland found that the human DNA repair enzyme (AGT) can specifically and rapidly react with benzylguanine (BG) derivatives. , combined by covalent bonds. Subsequently, the AGT was mutated to obtain a protein Snap-tag that does not react with guanine on the DNA strand. The protein is no longer localized in the nucleus, but has a large affinity with the small molecule BG derivative. Improve now [1-2]. The SNAP-tag is only 20KDa in size, so the target protein can be expressed in fusion with SNAP-tag. When BG with fluorescent probe label is introduced into the cell, BG and SNAP-tag will rapidly react covalently to achieve fluorescent labeling of the specific target protein in living cells (Fig. 1). Scientists can specifically select cell membrane permeable or non-permeable BG substrates for the study of membrane surface molecules and cytoplasmic expression of molecules [3]. Compared with fluorescent protein expression, the biggest advantage of SNAP-tag labeling is the flexible selectivity of fluorescent probes, such as fluorescent dyes, quantum dots, gold particles, and photoactivated, light-converted dyes. Currently, companies such as NEB have already adopted SNAP- The expression vector of the tag and these substrates are widely commercialized, and the spectral selection of the fluorescent probe is also very flexible (Table 1). Due to its ease of use, SNAP-tag has been widely used, such as to study the half-life, transport and interaction of proteins [4-6]. By transforming the SNAP-tag, the Johnsson team obtained a new CLIP-tag that was genetically engineered to not bind to the benzylguanine derivative, but to the benzylcytosine derivative (BC). Combine [7]. The combination of CLIP-tag and SNAP-tag enables the simultaneous labeling of two different proteins in the same cell, further extending the use of this tag self-labeling labeling method [8-11] (Table 1). In practice, the researchers added fluorescently labeled substrates to cells expressing SNAP-tag or CLIP-tag, and washed them several times to remove unbound fluorescent substrates for imaging, which is very simple and convenient. Table 1. A wide variety of SNAP-tag and CLIP-tag substrates currently available from NEB Application of SNAP-tag Technology in STED Ultra-High Resolution Microscopy In the past ten years, the microscopic imaging technology has developed rapidly, filling the optical microscope (~200 nm) to the electron microscope (~0.1 nm) resolution gap, and the ultra-high resolution microscope that breaks the optical diffraction limit is also becoming more and more popular. Matured. Among them, Professor Stefan Hell of the Max Planck Institute of Germany won the 2014 Nobel Prize in Chemistry for his research on stimulated emission depletion (STED) technology. The STED ultra-high resolution microscope is based on a confocal microscope, so its imaging speed is very fast and can be widely applied to ultra-high resolution imaging of living cells. In addition to traditional YFP and other fluorescent proteins can be used for ultra-high resolution imaging of STED living cells, SNAP-tag and CLIP-tag can easily introduce organic dyes such as AlexaFluo into living cells to achieve multi-color ultra-high resolution in living cells. Imaging. Organic dyes have better light stability, and the choice of spectrum is more flexible. With the specificity and stability of SNAP-tag and CLIP-tag markers, it can better serve long-term living cell ultra-high resolution microscopy. Imaging. Scientists such as Joerg B used SNAP-tag/BG-94 and CLIP-tag/BC-647 to label epidermal growth factor receptor (EGFR) and epidermal growth factor (EGF), respectively (Figure 2), using STED ultra-high resolution microscopy. The interaction between the two in living cells was analyzed (Fig. 3) [12]. Figure 2. Schematic representation of EGFR and EGF for SNAP-tag/BG-494 and CLIP-tag/BC-647N, respectively. In principle, homologous dimers are formed when EGFR and EGF are combined. Here, considering the simplicity and clarity of the schematic, EGFR is still expressed in monomeric form. references: 1. Juillerat, A., Gronemeyer, T., Keppler, A., Gendreizig, S., Pick, H., Vogel, H., and Johnsson, K. (2003). Directedevolution of O6-alkylguanine-DNA alkyltransferase for Efficient labeling of fusion proteins with small molecules in vivo. Chem Biol 10, 313-317. 2. Gronemeyer, T., Chidley, C., Juillerat, A., Heinis, C., and Johnsson, K. (2006). Directed evolution of O6-alkylguanine-DNA alkyltransferase for applications in protein labeling.Protein Eng Des Sel 19 , 309-316. 3.Brun, MA, Griss, R., Reymond, L., Tan, KT, Piguet, J., Peters, RJ, Vogel, H., and Johnsson, K. (2011). Semisynthesis of fluorescent metabolite sensors on cell Surfaces. J Am Chem Soc133, 16235-16242. 4. Banala, S., Maurel, D., Manley, S., and Johnsson, K. (2012). A caged, localizable rhodamine derivative for superresolution microscopy. ACS Chem Biol 7, 289-293. 5.Campos, C., Kamiya, M., Banala, S., Johnsson, K., and Gonzalez-Gaitan, M. (2011). Labelling cell structures and tracking cell lineage in zebrafish using SNAP-tag. Dev Dyn 240, 820-827. 6. Bannwarth, M., Correa, IR, Sztretye, M., Pouvreau, S., Fellay, C., Aebischer, A., Royer, L., Rois, E., and Johnsson, K. (2009). Indo-1 derivatives for local calcium sensing. ACS Chem Biol 4, 179-190. 7. Gautier, A., Juillerat, A., Heinis, C., Correa, IR, Jr., Kindermann, M., Beaufils, F., and Johnsson, K. (2008). Anengineered protein tag for multiprotein labeling in Living cells. Chem Biol 15, 128-136. 8. Maurel, D., Comps-Agrar, L., Brock, C., Rives, ML, Bourrier, E., Ayoub, MA, Bazin, H., Tinel, N., Durroux, T., Prezeau, L ., et al. (2008). Cell-surface protein-protein interaction analysiswith time-resolved FRET and snap-tag technologies: application to GPCRoligomerization. Nat Methods 5,561-567. 9.Comps-Agrar, L., Maurel, D., Rondard, P., Pin, JP, Trinquet, E., and Prezeau, L. (2011). Cell-surface protein-protein interaction analysis with time-resolved FRET and Snap-tag technologies: application to G protein-coupled receptor oligomerization. Methods Mol Biol 756, 201-214. 10. Feinstein, TN (2013). Cell-surface protein-protein interaction analysis with time-resolved FRET and snap-tagtechnologies. Methods Mol Biol 1066, 121-129. 11. Lukinavicius, G., Lavogina, D., Orpinell, M., Umezawa, K., Reymond, L., Garin, N., Gonczy, P., and Johnsson, K. (2013). Selective chemical crosslinking reveals a Cep57-Cep63-Cep152 centrosomalcomplex. Curr Biol 23, 265-270. 12. Pellett, PA, Sun, X., Gould, TJ, Rothman, JE, Xu, MQ, Correa, IR, Jr., and Bewersdorf, J. (2011). Two-color STED microscopy in living cells. Biomed Opt Express 2, 2364-2371. Concerned about Leica Micro WeChat: Chewable Tablets,Vitamin C Chewable Tablets,Chewable Vitamin C,Chewable Calcium Guangzhou Etechange Biological Engineering Co.,Ltd. , https://www.etechange.com